The Veg-X exchange standard
IAVS Ecoinformatics Working Group
2022-09-12
VegXStandard.RmdWhy a vegetation data exchange standard?
A primary technical impediment to large-scale sharing of vegetation data is the lack of a recognized international exchange standard for linking the panoply of tools and database implementations that exist among various organizations and individuals participating in vegetation research. In the absence of an exchange standard, the need for multiple, ad hoc mappings among databases and applications discourages merging of data and slows development of new analytical tools (Fig. 1a). By contrast, widespread use of a common exchange standard would avoid the need to repeatedly map data for synthetic projects by requiring only a single mapping between a given database or tool and the standard (Fig. 1b), thus facilitating data exchange and analysis. Application of an international exchange standard for vegetation data would form a critical part of the necessary infrastructure to allow these data to be combined for synthetic analysis at local and global scales.
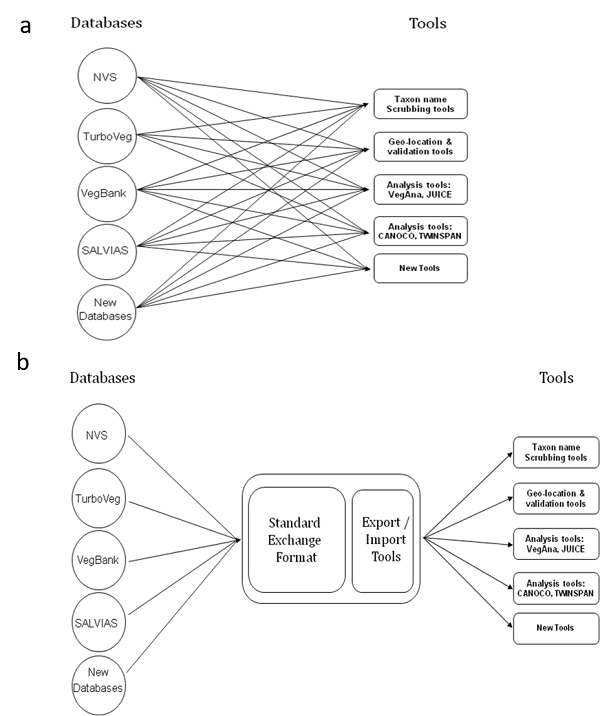
Fig. 1. Schematic diagram showing (a) problem: multiple tools and databases and (b) solution: sharing tools through a common standard.
Overview of the Veg-X standard
The Veg-X exchange standard for plot-based vegetation data (Wiser et al. 2011) is intended to be used to share and merge vegetation-plot data of different kinds. Veg-X allows for observations of vegetation at both individual plant and aggregated observation levels. It ensures that observations are fixed to physical sample plots at specific points in space and time, and makes a distinction between the entity of interest (e.g. an individual tree) and the observational act (i.e. a measurement). The standard supports repeated measurements of both individual organisms and plots, and enables the connection between the entity observed and the taxonomic concept associated with that observation to be maintained.
A goal in the creation of Veg-X was to have a schema that is relatively simple to read and use. To achieve this, highly nested structures were avoided and major vegetation data components were included (e.g. plot attributes, plot observations, organisms) as top-level elements that are referenced by each other through unique identifiers (e.g. a unique numerical ID) that allow the integrity of the original linkage to be captured. Although the main logical structure of vegetation data (i.e. the logical relationships between major data components) is fixed, alternative, user-defined ways of grouping observations are also allowed. As such, the standard can accommodate projects that are linked across time as well as longitudinal measures of plots or individuals to the extent that these are referenced in the original dataset through appropriate unique identifiers in those original sources.
The standard accommodates different data collection protocols by allowing specific aspects of data collection methods to be captured, such as whether plots were located subjectively or randomly, plot dimensions, definitions of cover-abundance scales, references to published measurement methods, etc. The standard also allows for the original units of measurement to be retained. All elements in the standard are clearly defined. This allows synonymous terms in source datasets to be mapped to a common set of concepts, thus overcoming the problems caused by inconsistent terminologies.
The plotObservation is the central Veg-X element, resulting from sampling a physical plot at a specific point in time, and can be related to one or more research projects (Fig. 2). The information about a sampled plot that is fixed over time (e.g. altitude, plot identifier or name, dimensions, aspect, slope, geology) and references to related plots (e.g. a parent plot) are stored in the separate element plot. By structuring the plot data in this way, repeat measures and nested plots can be accommodated in the standard.
Specific observations, either biotic or abiotic, are linked to the plot observation event. The standard allows storing observations of vegetation made at four different levels:
- Individual organism level, such as tree diameter (element individualOrganismObservation)
- Aggregate organism level (i.e. characteristic of a group of individuals taken together), such as the percent cover of a taxon (element aggregateOrganismObservation).
- Stratum level, such as the basal area of trees of the emergent layer (element stratumObservation).
- Community level, such as the successional stage or species richness (element communityObservation).
The standard maintains a clear distinction between the entity of interest (e.g. an individual organism, plot, or stratum) and the observation act (e.g. a measuring event applied to it). Together with unique identifiers that maintain the integrity of references between individual records within each component (e.g. between a plot and all the measuring events applied to it), the separation of components allows the standard to store multiple observations of the same entity (e.g. a plot or a tree). Analogously, a single observation event (e.g. a plot observation) may apply to multiple entities, thereby providing explicit grouping of entity observations. Each entity of interest (e.g. a tree) may have multiple observed properties (e.g. height, dbh) whose values are determined through measurement using a specific procedure or a method belonging to a particular protocol. Unlike individual organism observations, aggregate (i.e. collective) organism observations do not relate to a specific physical entity but provide estimates of the importance of a (abstract) taxonomic entity within the plot, such as through a cover estimate. Strata can be the subject of stratum observations (e.g. % of tree cover, tree height) and can be linked to aggregate organisms observations (Fig. 2).
The standard also maintains a distinction between identity of organisms (the taxon or taxon concept) and how these identities are applied to particular observations of organisms. This is done through three top-level elements:
- Element taxonConcept: A specific taxonomic concept (i.e. a name–reference combination).
- Element organismIdentity: The container for all information about the identity of an organism found in the plot observation (typically, this information includes a taxon name string possibly attached to taxonomic concept, but in some instances it may simply be the name of a morphospecies or a ‘field name’ used by the author of the data set). This element was called taxonNameUsageConcept in Wiser et al. (2011) and earlier versions of Veg-X.
- Element taxonDetermination: An assertion, made by a party, linking one or more taxonConcepts to a organismIdentity, normally after re-examining the herbarium voucher of the specimen.
All the organism observations referencing a given organismIdentity are affected by nomenclatural changes or determination events applied to it. This allows different determinations and taxonomic concepts to be associated with a vegetation entity so temporal changes in opinion regarding identification (i.e. “determination history”) can be recorded and both formal (i.e. taxon names) and informal (e.g. “field names”, “morphospecies”) names applied to a particular organism observation can be preserved. The fact that the organismIdentity is not nested within observations permits the same identity (i.e. name) to be reused within the scope of the individual dataset. Community determinations are handled in a similar way: communityDetermination elements allow a given plot observation to be related to one or multiple community concepts. Although the standard supports fully-specified taxonomic concepts, it does not require them. This is important as the full concept is unspecified and furthermore unrecoverable for most legacy data. On the other hand, because the schema can accommodate determination information (who did the identification, when, and with what reference), in theory it could be possible to recover concepts for many legacy datasets – in particular, tropical forest plots where such information is commonly preserved in the form of herbarium voucher specimens.
Veg-X is written as an XML schema, which is a definition of user-defined tags to structure textual information in order to create self-describing datasets. XML (Extensible Markup Language) is an open standard, and XML files are both machine and human-readable (they are stored in plain-text ASCII format). These characteristics help to ensure that data in this format will be accessible in the future. We made use of existing XML schema definitions, which we incorporated as modules of our schema. Specifically, we adapted parts of the Ecological Metadata Language (EML; https://knb.ecoinformatics.org/#tools/eml; Jones et al. 2006) to define entities like projects, protocols, parties and methods. To specify taxon concepts, we used element names adopted from the Taxon Concept Transfer Schema (TCS; http://www.tdwg.org/standards/117/). TCS can be used to support taxon concept mappings (e.g. Franz & Peet 2009).
The VegX R package
A barrier to the use of a standard like Veg-X is its complexity, which is nevertheless required to accommodate the wide variety of schemes existing for vegetation plot data sources. The large data integration projects employ eco-informaticians to achieve this, but the tools developed are specific to these projects and cannot be readily picked up by others. To make the exchange schema of Veg-X usable by the wider community requires the development of informatics tools for mapping data from different input formats (e.g. relevé tables from different databases, forest inventory data or stem-mapped forest plots) into Veg-X, mechanisms to create unique identifiers to allow source datasets to be combined, and tools to export data from Veg-X to a range of formats that can serve as input to software packages for data analysis and visualisation.
In 2017, the Ecoinformatics Working Group of the International Association for Vegetation Science (IAVS; http://iavs.org/Working-Groups/Ecoinformatics.aspx ) decided to develop an R package to promote the usage of the Veg-X standard. The R package is specifically intended to be used to:
- Integrate and harmonize vegetation-plot data from different sources with the aim of conducting new analyses.
- Produce archivable XML documents for vegetation-plot that have no ‘home’ in existing vegetation plot databases such as those catalogued by the Global Index of Vegetation Plot Databases (GIVD; www.givd.info ) or as needed to support data availability requirements to support scientific publications.
The development of VegX R package has been conducted in parallel to an extensive revision of Veg-X, which has led to the deployment of version 2.0 of the exchange standard. Moreover, the package does not currently include all the main elements and sub-elements of Veg-X (see next section). This was a practical decision to enable a usable tool to be developed to meet the purpose described above and not be overly complex for users. Future versions of the package may allow more elements of the standard to be used while accounting for backward compatibility with Veg-X files conforming to version 2.0 or later versions.
Data structure of Veg-X (ver. 2.0)
Veg-X has a non-hierarchical data structure. Different data elements relate to each other via identifiers in a flexible way. The following figure illustrates the relationships between the main elements of Veg-X.
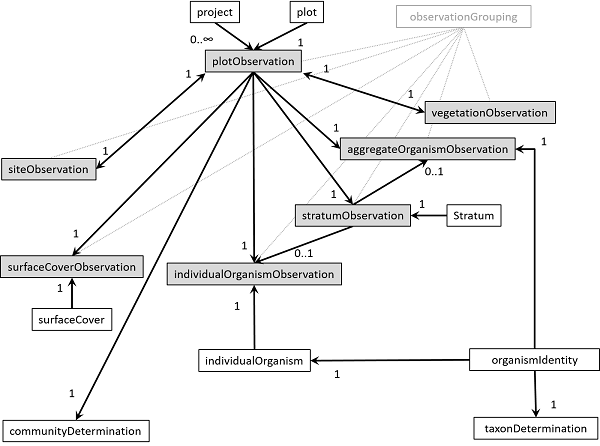
Fig. 2. Main VegX elements and their logical relationships. Arrows indicate that an identifier of the origin element is referenced in the destination element. Accompanying numbers indicate the number of instances of the origin element that are allowed to be referenced in the destination element. Observations are in tinted boxes.
The following table provides the brief descriptions of the main elements of Veg-X (column ‘R’ indicates whether the sub-elements are currently implemented in the VegX R package):
| Main element | Description | R |
|---|---|---|
| project | Describes the research context in which the dataset was created, including descriptions of over-all motivations and goals, funding, personnel, description of the study area etc. | Yes |
| plot | A plot is a sampling location. It records the properties of the vegetation plot independent of time and can be referenced by many observations by links to the unique plot code. | Yes |
| plotObservation | Observations made on a single plot and during a single date-time period. This allows all time dependent parameters to be grouped. | Yes |
| organismIdentity | The identity of an organism occurring within the dataset. This is a name defined by the dataset author which may follow, or not, nomenclatural codes. It may be further related to a published name (taxon name, tcs:TaxonName) and or taxonomic name (taxon concept, tcs:TaxonConcept) in taxonDetermination. | Yes |
| individualOrganismObservation | An observation applying to one occurrence of an organism (or part of an organism). It is a container for measurements made on the organism (e.g. diameter, height, crown dimensions, biomass, growth form, number of stems). | Yes |
| individualOrganism | An identified organism recorded during one or more individual organism observation events. Individuals may have an identification label (e.g. tree tag number). | Yes |
| aggregatedOrganismObservation | An observation applying to all occurrences of an organism based on an aggregation factor. | |
| stratumObservation | A specific observation applying to a stratum in a single plot during a single date-time period. Each stratum measurement may be referenced by observations of taxa within a plot. For example, abundance estimates of a taxa on a plot within a specific stratum. | Yes |
| stratum | The specific definition of a stratum referred to by observations in the dataset. A stratum usually belongs to a ordered list that together are the set of strata definitions in use in a specific dataset. | Yes |
| communityObservation | A container for measurements that apply to the entire plant community and made on a single plot during a single date-time period. | Yes |
| siteObservation | A container for all the site (i.e. soil, climate, landuse, habitat, …) measurements made on a single plot during a single date-time period. Unlike other observation elements, it relates to plotObservation in a one-to-one relationship. | Yes |
| surfaceType | The definition of a surface type, not the observation of cover on it. | Yes |
| surfaceCoverObservation | A single cover measurement applying to a surface type in a single plot during a single date-time period. | Yes |
| taxonDetermination | A specific relationship or assertion between two name concepts which are not part of the original definition of either of these concepts; possibly by a third party. This typically allows for an organism identity to be linked to a specific taxa treatment (taxon concept), according to a third party. Similar to a tcs TaxonRelationshipAssertion. | No |
| communityDetermination | An identification applying one or more community concepts to a plot observation by a party. | No |
| observationGrouping | An specific grouping of observation records, of any kind, that are grouped in the data management system owing to some common characteristic. | No |
Additional resource elements of the Veg-X schema are used to provide information about people, methods, bibliographic references, organism names and concepts:
| Resource element | Description | R |
|---|---|---|
| party | Describes a responsible party (person or organization), and is typically used to name the originator of a resource or metadata document. | Yes |
| attribute | A specific definition of a measured property. An attribute has to be one of three types: qualitative (unordered categorical variable, i.e. nominal), ordinal (ordered list of values) or quantitative (a numerical variable, either discrete or continuous. | Yes |
| method | A specific method definition followed in the creation of the dataset (e.g. the measurement of pH, the estimation of plant abundance or the definition of strata). | Yes |
| protocol | A specific grouping of methods related by common action. A protocol may have many method or steps. | No |
| literatureCitation | Provides overview information about the literature, including citation string and DOI. | Yes |
| organismName | The name of an organism used in the data set. This will normally be a nomenclatural unit of any rank (order, family, genus, species, subspecies, etc.). If it is a formal scientific name (not necessarily including authority) then the attribute ‘taxonName’ should be set to true. However, the organism name can be a morphospecies, a field name… cases in which the attribute ‘taxonName’ should be set to false. | Yes |
| taxonConcept | Representation of a taxon concept (i.e., an organism name and the organism description given by an author in a publication). A taxon concept may be referenced in an organism identity as the original concept used by the author of the data set, or it can be referenced in taxonDetermination allowing an organism identity to be mapped to a taxonomic concept by third parties after re-examination. | Yes |
| communityConcept | A name and some kind of definition of a community type, preferably a community name as used in a reference. | No |
Details of the main Veg-X elements
In the following sections we detail the sub-elements of all main elements (except communityDetermination and taxonDetermination) in the Veg-X standard, along with some instructions on how the standard should be used. The VegX package maintains unique IDs for all main elements, for internal consistency. However, these IDs are largely hidden from the user (but you will see ID references in the descriptions below). From each element described, we indicate the combination of fields that uniquely identify them and are used to guide element merge. In the tables describing sub-elements, we use column ‘#’ to indicate the number of times they can (or must) occur to have a valid Veg-X document (‘1’ means they occur one and are required; ‘0..1’ means they are optional and can occur only once; ‘0..n’ means they are optional and can occur many times; ‘1..n’ means they must occur at least once, but can occur many times). As before, column ‘R’ (‘Yes’/‘No’/‘Partial’) indicates whether the sub-elements are currently implemented in the VegX R package (‘Partial’ indicates an implementation that is not complete).
Element project
A project element describes the research context in which the dataset was created, including descriptions of over-all motivations and goals, funding, personnel, description of the study area etc. The definition of Veg-X project elements was borrowed from the Ecological Metadata Language (v. 2.0.1). from the user’s perspective projects are uniquely identified by their sub-element title.
| Sub-element | Description | # | R |
|---|---|---|---|
| title | Title of the project. | 1..n | Yes |
| personnel | Contact and role information for people involved in the research project. | 1..n | Yes |
| abstract | A brief description of the aims and findings of the project. | 0..1 | Yes |
| funding | Funding information. | 0..1 | Yes |
| studyAreaDescription | Description of the physical area associated with the research project, potentially including coverage, climate, geology, distrubances, etc. | 0..1 | Yes |
| designDescription | Description of the design of the research project (specially overall plot placement). | 0..1 | Yes |
| relatedProjectID | A link to another project, by ID. | 0..n | No |
| documentCitationID | A link, by ID, to the citation of a document describing the project. | 0..n | Yes |
Element plot
A plot is a sampling location, represented as one or more points, lines, polygons, or volumes, and is the basis for experimentation or measurement. Its properties are assumed to be constant over time. A point within the plot may be used as center for relative coordinates, which are required to be Cartesian. Plots may have no explicit bounds, and may refer to an area of inference. A plot may be related to other plots in order to express parent-child, contiguity, or other type of relationship.
The element plot records the properties of the vegetation plot that are independent of time. The Veg-X standard allows storing globally unique identifiers for plot elements, but the VegX package uses its own set of IDs for internal consistency. From the perspective of the provider of a single data set, plots can be uniquely identified by their sub-element plotName. However, when merging data sets plotUniqueIdentifier from different sources, helps distinguishing plots that may have the same name but come from different sampled areas. Even though the standard allows different kinds of spatial relationships between plots to be specified, currently the R package only enables parent-child relationships to be specified.
| Sub-element | Description | # | R |
|---|---|---|---|
| plotName | Name or label for a plot, unique within the data set | 1 | Yes |
| plotUniqueIdentifier | Plot identifier that is unique across the dataset, derived from the data source, and preferably globally unique. | 0..1 | Yes |
| relatedPlot | A plot may be related spatially to other plots in order to express parent-child, sub-plot or contiguity. | 0..n | Partial |
| placementMethod | Strategy followed when placing this particular plot. Useful for example if different sampling strategies have been followed within one project. | 0..1 | No |
| placementPartyID | A link to a party that participated in the establishment of the plot, by ID. | 0..n | Yes |
| placementNote | Additional comments or explanations regarding plot placement. | 0..n | No |
| location | Information regarding the location of the plot on earth’s surface. | 0..1 | Partial |
| geometry | Information regarding the geometry of the plot (area, shape, dimensions, coordinates, …) as well as the point within the plot that serves as the plot origin for location. | 0..1 | Partial |
| topography | Information regarding the shape and features of the surface on which the plot was placed (e.g. aspect, slope, …). | 0..1 | Yes |
| parentMaterial | Underlying geological material (generally bedrock or a superficial or drift deposit) in which soil horizons form. | 0..n | No |
Sub-elements location and geometry are specially important, as they contain the description of plot location and shape, respectively. For this reason we describe these child elements in some detail.
Sub-element location
Location stores information regarding the location of a plot on the earth surface. Child elements horizontalCoordinates are used to store x-y coordinates in a spatial reference system. To avoid ambiguity there should be only one coordinate pair for a plot, and the implementation of the VegX R package follows this rule. However, the Veg-X schema can accomodate multiple coordinate measurements made by different parties or at different times. The same applies to verticalCoordinates, locationInWaterBody and gridPosition (the later two not covered in the R package).
| Sub-element | Description | # | R |
|---|---|---|---|
| horizontalCoordinates | Horizontal coordinates of a plot on the Earth’s surface (i.e. x-y coordinates in a spatial reference system). | 0..n | Partial |
| verticalCoordinates | Elevation of the plot in respect to some vertical datum (such as the mean sea level or an elliposid). | 0..n | Partial |
| markers | Information about markers (like magnetic markers or wooden pegs) that help locating the plot. There should also be a description about where in the plot the markers are found (for example in the corners or in the centre). | 0..1 | No |
| locationInWaterBody | Location in respect to a water surface or shoreline. | 0..n | No |
| gridPosition | Position in a grid such as used for floristic surveys. | 0..n | No |
| authorLocation | Descriptive note about the original location described by author. | 0..1 | No |
| locationNarrative | Text description that provides information useful for plot relocation. | 0..1 | No |
| places | A collection of named places or geographic regions. Includes elements to indicate what type of place and which place/geo-region schema it was from. | 0..n | Yes |
Sub-element geometry
Sub-element geometry stores information regarding the geometry of the plot (area, shape, dimensions, coordinates, …) as well as the point within the plot that serves as plot origin for location. Sub-element geometry may be lacking in plot-less vegetation observations. When present, the user of the schema has to choose one plot shape: circle, rectangle, line or polygon. Currently, the VegX R package allows storing information about area, shape and dimensions, but not plot origin, orientation and path.
| Sub-element | Description | # | R |
|---|---|---|---|
| area | Total area of the plot. Usually recorded in square meters. | 0..1 | Yes |
| shape | Plot’s shape: linear, rectangle, polygon or circle. | 0..1 | Yes |
| plotOrigin | Definition of the position of the plot origin within the plot (here usually “center”). This is referred to in the horizontalCoordinates element under locationInPlot. The actual coordinates go into the location - horizontalCoordinates element | 0..1 | No |
| radius | Define the radius of circular plots. Usually recorded in meters. | 0..1 | Yes |
| width | Width of a regular rectangle. In case of a square plot, the width of both sides. | 0..1 | Yes |
| length | Length (largest dimension) of a rectangle plot or length of a linear plot. | 0..1 | Yes |
| orientation | Orientation of the main axis of the plot (e.g. in degrees from North). For quadrat plots the axis closer to the N-S axis should be given. | 0..1 | No |
| bandWidth | Distance from the linear plot axis. This distance delimits the surface included for measurements. | 0..1 | Yes |
| path | Set of points conforming the path in a linear plot (i.e. a transect) | 0..1 | No |
| outerBoundary | Absolute or relative coordinates defining the outline of a polygon. | 0..1 | No |
| innerBoundary | Coordinates defining any inner boundary of a polygon | 0..1 | No |
Element plotObservation
An element plotObservation is used to group all observations made on a single plot and during a single date-time period. While the Veg-X standard allows globally unique identifiers for plotObservation elements to be stored, the VegX package uses its own set of IDs for internal consistency. From the user’s perspective, plot observations are uniquely identified by the plot’s name (and its unique identifier, if present) and obsStartDate.
While the schema allows multiple references to project elements, the VegX package only enables a single reference to a project element to be specified (sub-element projectID). Similarly, the package allows only one party to be specified, among those involved in the plot survey (sub-element observationPartyID).
| Sub-element | Description | # | R |
|---|---|---|---|
| plotID | A link to a specific plot by the plot’s ID . | 1 | Yes |
| obsStartDate | The start date of this specific observation of the plot. Recorded in ISO 8601 date format: yyyy-mm-dd. | 1 | Yes |
| obsEndDate | The end date of this specific observation of the plot. Recorded in ISO 8601 date format: yyyy-mm-dd. | 0..1 | Yes |
| plotObservationUniqueIdentifier | Plot observation identifier that is unique across the dataset, derived from the data source, and preferably globally unique. | 0..1 | Yes |
| projectID | A link to a specific ‘project’ by ID. | 0..n | Partial |
| previousObservationID | A link to previous plot observations. | 0..n | No |
| communityObservationID | A link to a specific community observation by ID. Note that the relationship is one-to-one. Only one community observation (with potentially many measurements inside) is allowed for each plot observation. | 0..1 | Yes |
| siteObservationID | A link to a specific site observation by ID. Note that the relationship is one-to-one. Only one site observation (with potentially many measurements inside) is allowed for each plot observation. | 0..1 | Yes |
| previousObservationID | A link to a previous plot observation, by ID. Not normally necessary as observations can be ordered via obsStartDate. | 0..n | No |
| observationPartyID | A link to a party that participated in the observation of the plot, by ID. | 0..n | Partial |
| license | License linked to this plot observation. | 0..1 | No |
| taxonomicQuality | Subjective assessment of the taxonomic quality on the plot. | 0..n | No |
| observationNarrative | Additional unstructured observations useful for understanding the ecological attributes and significance of the plot observations. | 0..1 | No |
| observationConditions | Conditions at the time of observation. | 0..1 | No |
| referencePublication | Reference to an original publication and additionally a table or section within a publication. | 0..n | No |
| observationGroupingID | A reference to a specific observation grouping by ID. | 0..n | No |
| observationNote | Additional comments or explanations pertaining to the observation event. | 0..n | No |
Element organismIdentity
The identity of an organism (or a set of organisms) occurring within the dataset. This is initially a name defined by the dataset author which may or not be following nomenclatural codes. The identity may be complemented with an scientific name (taxon name) accepted according to a specified nomenclatural authority, by using the sub-element preferredTaxonNomenclature. The taxonomic concept (taxon name + reference defining the concept) that the author of the observation had in mind when observing the organism (or that a third party assumes he had in mind) can also be specified in the sub-element originalIdentificationConcept. Subsequent re-evaluations of the taxon concept (e.g. after inspection of the herbarium voucher) by third parties should be specified using the main element taxonDetermination (which are not currently supported by the VegX package).
| Sub-element | Description | # | R |
|---|---|---|---|
| originalOrganismNameID | A link, by ID, to an organism name (e.g. normally a taxon name, but not necessarily including the authority, or even field names, morphospecies, …) that the author of the dataset originally used to refer to an organism observed within the plot. The taxon names used as label the organism identity should not contain spelling errors, but they may not be the accepted name according to current nomenclature codes. | 1 | Yes |
| originalIdentificationPartyID | A link to a party involved in the original organism identification (normally the author of the data set), by ID. | 0..n | No |
| originalIdentificationNote | Additional comments or explanations pertaining to the original identification of the organism. | 0..n | No |
| voucher | Herbarium accession number for any archived voucher specimens. | 0..n | No |
| originalIdentificationConcept | The taxon concept originally associated to the organism identity. This may have been specified by the author of the data set, or it may be asserted by a third party based on information such as date of observation or geographic location. | 0..n | Partial |
| preferredTaxonNomenclature | The interpretation of the nomenclature that should be applied to organism identity, made after the observation event by the author of the data set or a third party. The sub-element preferredTaxonNameID points to an organism name that is the accepted name according to the current nomenclature. | 0..1 | Partial |
When displaying organism observations, the VegX package uses a field called organismIdentityName to name organisms with the following rules:
- If preferredTaxonNomenclature has NOT been defined then organismIdentityName is equal to the originalOrganismName (via originalOrganismNameID).
- If preferredTaxonNomenclature has been defined then organismIdentityName is equal to the preferredTaxonName (via preferredTaxonNameID).
Sub-element originalIdentificationConcept
Stores the taxon concept originally associated to the organism identity. This may have been specified by the author of the data set, or it may be asserted by a third party based on information such as date of observation or geographic location.
| Sub-element | Description | # | R |
|---|---|---|---|
| taxonConceptID | A link to the taxon concept stated by the author, or as asserted by a third party based on information such as the date of the observation, geographic location, etc. | 1 | Yes |
| conceptAssertionDate | Date of the taxon concept assertion. Recorded in ISO 8601 date format: yyyy-mm-dd. | 0..1 | Yes |
| conceptAssertionPartyID | A link, by ID, to a party involved in the assertion of the original taxon concept. | 0..n | Partial |
| conceptAssertionNote | Additional comments or explanations pertaining to the assertion of the original taxon concept. | 0..n | No |
Sub-element preferredTaxonNomenclature
Stores the interpretation of the nomenclature that should be applied to organism identity, made after the observation event by the author of the data set or a third party.
| Sub-element | Description | # | R |
|---|---|---|---|
| preferredTaxonNameID | A link to a scientific taxon name (i.e. an organism name whose attribute ‘taxonName’ is true) accepted to label the organism identity appropriately, as stated by the author of the data set or a third party responsible for its nomenclature. | 1 | Yes |
| interpretationDate | Date for the last nomenclature revision applied to this organism identity. Recorded in ISO 8601 date format: yyyy-mm-dd. | 0..1 | Yes |
| interpretationSource | A string describing the source for the last nomenclature interpretation applied to this organism identity (i.e. the Plant List). | 0..1 | Yes |
| interpretationCitationID | A link to the publication where nomenclature interpretation is described. | 0..1 | Yes |
| interpretationPartyID | A link to a party who undertook the nomenclarure revision. | 0..n | Partial |
| interpretationSource | Additional comments or explanations pertaining to the nomenclature interpretation. | 0..n | Np |
Element stratum
The definition of a stratum, not the observation of a stratum. An individual stratum usually belongs to a ordered list that together are the set of strata definitions in use within a specific dataset. This set of strata will normally have been defined according to the same method, but individual stratum may also be assigned a method. Strata that are defined from limits in a quantitative measurement, like height, the user of the Veg-X standard can use the method pointed by methodID to describe the quantitative attribute associated to the stratum definition (e.g. height in m). From the package user’s perspective, strata are uniquely identified by the name of the stratum definition method and the stratumName.
| Sub-element (quantitative) | Description | # | R |
|---|---|---|---|
| stratumName | Name associated with this stratum and which identifies it. | 1 | Yes |
| methodID | A reference to a specific method used to define this stratum. | 0..1 | Yes |
| definition | A longer description of the stratum definition. | 0..1 | Yes |
| order | An indication of a position in an ordered sequence of strata. | 0..1 | Yes |
| lowerLimit | Lower limit of the stratum in some known dimension (e.g. height) defined in the attribute of the method pointed to by ‘methodID’. | 0..1 | Yes |
| upperLimit | Upper limit of the stratum in some known dimension (e.g. height) defined in the attribute of the method pointed to by ‘methodID’. | 0..1 | Yes |
Element stratumObservation
A stratumObservation is a specific observation applying to a stratum in a single plot during a single date-time period. Each stratum observation may be referenced by observations of taxa or individuals within a plot. For example, abundance estimates of a taxa on a plot within a specific stratum. In addition, the stratumObservation may contain measurements of the lower and upper vertical limits of the stratum (if those are not fixed by the stratum definition) and an assessment of plant abundance like cover or number of individuals. A stratumObservation always contains a reference to a plotObservation, where contextual information lies (plot, project, parties, date-time period). It also contains a reference to a stratum, which contains its definition. From the package user’s perspective, stratum observations are uniquely identified by the name of the stratum definition method, the stratum name and the plot observation.
| Sub-element | Description | # | R |
|---|---|---|---|
| stratumID | A reference to a specific stratum by ID. | 1 | Yes |
| plotObservationID | A reference to a specific plotObservation. | 1 | Yes |
| lowerLimitMeasurement | A measurement of the lower limit (i.e. height) of the stratum. | 0..1 | Yes |
| upperLimitMeasurement | A measurement of the upper limit (i.e. height) of the stratum. | 0..1 | Yes |
| stratumMeasurement | A measurement (e.g. plant cover, or individual count) made in the stratum. | 0..n | Yes |
| observationGroupingID | A reference to a specific observation grouping by ID. | 0..n | No |
| observationNote | Additional comments or explanations regarding this observation. | 0..n | No |
Element aggregateOrganismObservation
An aggregateOrganismObservation is an observation applying to all occurrences of an organism (e.g. a taxon). An aggregateOrganismObservation contains a reference to a single plotObservation and a link to a organismIdentityID, which can be linked to all the taxon identification information. Optionally, it may also link to a stratumObservation. It may contain one of several sub-elements aggregateMeasurement, each of them being an assessment of the overall occurrence of an organism in a Plot (e.g. number of stems, percentage cover, total biomass, basal area). If there is no instance of aggregateMeasurement, then the taxon is understood to be simply present. From package user’s perspective, aggregate organism observations are uniquely identified by plot observation and organism identity, and by the stratum observation when defined.
| Sub-element | Description | # | R |
|---|---|---|---|
| plotObservationID | A link to a specific plot observation by ID. | 1 | Yes |
| organismIdentityID | A link to a specific organism identity by ID. | 1 | Yes |
| aggregateOrganismMeasurement | A measurement for a aggregate organism value (e.g. plant cover of a taxon). Values can be further defined uppervalue, accuracy etc. Many measurements (e.g. counts, cover, basal area…) can be added to the same aggregate organism observation. | 0..n | Yes |
| heightMeasurement | Optional height at which the aggregated observation was made, e.g. in meters. It applies to all aggregate measurements included in this aggregateOrganismObservation. | 0..1 | Yes |
| stratumObservationID | A link to a specific stratumObservation by ID. It applies to all aggregate measurements included in this aggregateOrganismObservation. | 0..1 | Yes |
| observationGroupingID | A link to a specific observation grouping by ID. | 0..n | No |
| observationNote | Additional comments or explanations regarding this observation. | 0..n | No |
Element individualOrganism
An element individualOrganism represents an organism recorded during one or more observation events and identified through an identification label (e.g. tree tag number). In Veg-X documents, individual organisms may or may not have been given a taxon name (i.e. a link via sub-element organismIdentityID), and the standard allows specifying the relative position of individuals within the plot to which they belong, as well as to specify related individuals. From the perspective of the VegX package user, individual organisms are identified by plot identity (i.e. name and unique identifier if defined) and the label of the individual organism.
| Sub-element | Description | # | R |
|---|---|---|---|
| plotID | A reference to a specific plot by the plot ID. | 1 | Yes |
| individualOrganismLabel | A label that is associated with an individual (e.g. a numerical tree tag). | 1 | Yes |
| organismIdentityID | A reference to a specific organismIdentity by ID. | 0..1 | Yes |
| birthDate | Date of birth recorded in ISO 8601 date format: yyyy-mm-dd. | 0..1 | No |
| relatedIndividual | An item may be related or connected in some way to other items. For example fused stems or epiphytic relationships. | 0..n | No |
| location | Information regarding the location of an organism on the earth’s surface, either absolute or relative to the plot origin or to a related individual. | 0..1 | No |
| individualOrganismNote | For specifying additional comments or explanations pertaining to the individual organism. | 0..n | No |
While relatedIndividual is used to link to other organisms in an arbitrary relationship (e.g. epiphytes or fused stems), quantitative spatial relationships should be specified in location, as discussed below.
Sub-element location
The location of an organism is assumed to be constant in time during the organism lifespan. Sub-elements horizontalLocation are used to store x-y (or polar) coordinates of organism, either absolute or in relation to the plot origin or a related organism. To avoid ambiguity there should be only one coordinate pair for an organism. However, the Veg-X schema can accomodate multiple measurements made by different parties or at different times. The R package does not yet support location elements for organisms.
| Sub-element | Description | # | R |
|---|---|---|---|
| horizontalLocation | Horizontal location of an organism on the Earth’s surface (absolute or relative to a reference point such as the plot origin or a related individual). | 0..n | No |
| verticalLocation | Elevation of the item in respect to some vertical datum (such as the mean sea level or an ellipsoid) or in relation to the plot origin or to a relatedIndividual. | 0..n | No |
| markers | Information about markers (like tags) that help locating the organism. | 0..1 | No |
| quadrant | One out of four quadrats such as those used in Point-Centred Quarter Method (PCQM) | 0..n | No |
Element individualOrganismObservation
An element individualOrganismObservation is an observation applying to one occurrence of an organism (or part of an organism). It is a container for measurements made on the organism (e.g. diameter, height, crown dimensions, biomass, growth form, number of stems). An individualOrganismObservation contains a reference to a unique plotObservation and to an individualOrganism. Optionally, the individualOrganismObservation may link to a stratumObservation. Regarding measurements, the schema includes specific subelements to store the measurement of plant height and stem diameter, the latter including or not a measurement of distance from the ground at which diameter was measured. Other measurements such as growth form, canopy dimensions, health status… can be included in sub-elements individualOrganismMeasurement (for simple measurements) or individualOrganismMultipleMeasurement for multiple (tuple) measurements.
From the package user’s perspective, individual organism observations are uniquely identified by plot observation and the label of the individual organism.
| Sub-element | Description | # | R |
|---|---|---|---|
| plotObservationID | A reference to a specific plotObservation by ID. | 1 | Yes |
| individualOrganismID | A reference to a specific individualOrganism by ID. | 1 | Yes |
| stratumObservationID | A reference to a specific stratum observation that this individual was measured in on the plot. | 0..1 | Yes |
| heightMeasurement | Measurement of the maximum height reached by the observed individual. | 0..1 | Yes |
| diameterMeasurement | Diameter of the stem without explicit measurement of base distance (may be defined in the measurement method definition) | 0..1 | Yes |
| diameterBaseDistanceMeasurement | A container for diameter measurements at a given distance along the stem from the ground. | 0..1 | No |
| individualOrganismMeasurement | A measurement applying to the observed individual. This includes qualitative, ordinal or quantitative assessments of form, health, dimensions of components, … | 0..n | Yes |
| individualOrganismMultipleMeasurement | A n-tuple of a related measurements (e.g. paired data such as length, width and height to calculate volume). The definition of the relationship type is left open to the user, but is intended to allow specifying a set of measurements that have to be considered together. | 0..n | No |
| observationGroupingID | A reference to a specific observation grouping by ID. | 0..n | No |
| observationNote | Additional comments or explanations regarding this observation. | 0..n | No |
Element communityObservation
A communityObservation is a container for all measurements that apply to the entire plant community and are made on a single plot during a single date-time period. Unlike other observation elements, it relates to plotObservation in a one-to-one relationship. The reason is because, unlike other entities, there is a single entity to which measurements refer. There is no specific variable to identify uniquely community observations, but they are uniquely identified by their related plot observation.
| Sub-element | Description | # | R |
|---|---|---|---|
| plotObservationID | A reference to a specific plotObservation. | 1 | Yes |
| communityMeasurement | A measurement (e.g. number of individuals, basal area) applying to the whole plant community or forest stand. | 0..n | Yes |
| successionalType | Description of the assumed successional status of the plot. This description is of necessity highly subjective. | 0..n | No |
| observationGroupingID | A reference to a specific observation grouping by ID. | 0..n | No |
| observationNote | Additional comments or explanations regarding this observation. | 0..n | No |
Element siteObservation
A siteObservation is a container for all the site (i.e. soil, climate, landuse, habitat, …) measurements made on a single plot during a single date-time period. Unlike other observation elements, it relates to plotObservation in a one-to-one relationship. The reason is because, unlike other entities, there is a single entity (i.e. the site) to which all measurements refer. There is no specific variable to identify uniquely site observations, but they are uniquely identified by their related plot observation. Abiotic (i.e. soil, climate or water body) measurements are a special kind of measurements within the Veg-X schema, in the sense that: (1) they can have IDs and can thus be related to each other: (2) and they can have relative coordinates of the measurement within the plot, in the same way as individuals.
| Sub-element | Description | # | R |
|---|---|---|---|
| plotObservationID | A reference to a specific plotObservation. | 1 | Yes |
| soilMeasurement | A measurement of a soil attribute (soil chemistry, soil texture, structure, …). | 0..n | Yes |
| climateMeasurement | A measurement of a climate attribute. | 0..n | Yes |
| waterBodyMeasurement | A measurement of an attributes of a water body within the plot (e.g. water level, not for soil water, which should be included in soilMeasurement). | 0..n | Yes |
| soilType | A specific soil type, applying to this plot during the plot observation. | 0..n | Yes |
| humusType | A specific humus type, applying to this plot during the plot observation. | 0..n | Yes |
| climateType | A specific climate type, applying to this plot during the plot observation. | 0..n | Yes |
| hydrologicRegimeType | Reflection of frequency and duration of water level variations, applying to this plot during the plot observation. | 0..n | Yes |
| legalProtection | Legal protection status of the plot during the plot observation. Recommended that this is from a closed list of legal protection status types. | 0..1 | No |
| landuse | A specific land use type, for example pasture, applying to this plot during the plot observation. | 0..n | No |
| habitat | A specific habitat type, applying to this plot during the plot observation. | 0..n | No |
| observationGroupingID | A reference to a specific observation grouping by ID. | 0..n | No |
| observationNote | Additional comments or explanations regarding this observation. | 0..n | No |
Element surfaceTypes
The definition of surface types, not the observation of cover on them. From the package user’s perspective, surface types are uniquely identified by the name of their definition method the surface type name.
| Sub-element (quantitative) | Description | # | R |
|---|---|---|---|
| surfaceName | Name associated with this surface type and which identifies it. | 1 | Yes |
| methodID | A reference to a specific method used to define this surface type. | 0..1 | Yes |
| definition | A longer description of the surface type definition. | 0..1 | Yes |
Element surfaceCoverObservation
A surfaceCoverObservation is a single cover measurement applying to a surface type in a single plot during a single date-time period. From the package user’s perspective, surface cover observations are uniquely identified by surface type and plot observation.
| Sub-element (quantitative) | Description | # | R |
|---|---|---|---|
| plotObservationID | A link to a specific plotObservation by ID. | 1 | Yes |
| surfaceTypeID | A link to a specific surface type by ID. | 1 | Yes |
| coverMeasurement | The cover measurement, usually in percent cover of the surface when projected to the ground. | 1 | Yes |
Element observationGrouping
A specific grouping of observation records, of any kind, that are grouped in the data management system owing to some common characteristic. For example, records that represent revisits to the same area for monitoring purposes can be linked together through this entity. Note that some specific groupings are already defined in the schema and therefore they should not be repeated (e.g. the grouping of observations made on a specific plot during a specific time is a plotObservation).
| Sub-element (quantitative) | Description | # | R |
|---|---|---|---|
| name | The unique name of a specific grouping entity that is subsequently referenced by specific observations. | 1 | No |
| type | The grouping entity type. For example, grouping of individual organism observations for the purposes of describing a physical relationship. Recommended that a closed-list is developed and used. | 1 | No |
Details of resource elements
In the following sections we detail the sub-elements of all resource elements (except protocol and communityConcept) in the Veg-X standard. From each element described, we indicate the combination of fields that uniquely identify them and are used to guide element merge.
Element party
A party element describes a responsible party (person, organization or a position), and is typically used to name the originator of a resource or metadata document. Parties are uniquely identified by the party name, which is either an individualName, organizationName or positionName.
| Sub-element | Description | # | R |
|---|---|---|---|
| individualName | The full name of the person being described. | 0..1 | Yes |
| organizationName | The full name of the organization being described. | 0..1 | Yes |
| positionName | The name of the title or position associated with the resource. | 0..1 | Yes |
| address | The full address information for a given responsible party entry. | 1..n | Yes |
| phone | Information about the contact’s telephone. | 0..1 | Yes |
| electronicMailAddress | The email address of the contact. | 0..1 | Yes |
| onlineURL | A link to associated online information, usually a web site. | 0..1 | Yes |
Element literatureCitation
An element literatureCitation provides information about a literature reference, including citation string and DOI.
| Sub-element | Description | # | R |
|---|---|---|---|
| citationString | A string indicating the citation reference | 0..1 | Yes |
| citationDOI | A string indicating the DOI that points to the resource. | 0..1 | Yes |
Element method
An element method provides the definition of a specific method followed in the creation of the dataset (e.g. the measurement of pH, the estimation of plant abundance or the definition of strata). From the perspective of the VegX package user, methods are identified uniquely through their element name. An important sub-element of a method is its subject, which contains the description of an attribute class to which the method applies, and is used for combining values which may be initially obtained using different methods. For example, subject would be pH measurement of upper soil solution, whereas a particular methods for this subject would be the measurement in water or measurement in 0.01 mol CaCl. All attributes pointing to a given method are assumed to apply to the same subject.
| Sub-element | Description | # | R |
|---|---|---|---|
| name | Name associated with the method. For example, “percent cover”. | 1 | Yes |
| description | A brief description of the method (e.g., measured parameter or basal area of all stems > 10 cm dbh or counts of all saplings >1.35 m tall and less than 2 cm dbh). | 1 | Yes |
| subject | The description of an attribute class for comparative purposes. If two methods measure the same attribute (e.g. plant cover), but with different degrees of precision and accuracy, setting ‘subject’ to ‘plant cover’ allows combining their values. All attributes pointing to the same method are assumed to apply to the same subject. | 1 | Yes |
| protocolID | A reference to a specific protocol by its ID. | 0..1 | No |
| citationID | A reference to a specific citation of literature, by ID, where the method is explained in length. | 0..1 | Yes |
Element attribute
An attribute element contains the definition of a specific measured property. An attribute has to be one of three types: qualitative (unordered categorical variable, i.e. nominal), ordinal (ordered list of values) or quantitative (a numerical variable, either discrete or continuous. The sub-elements of attributes depend on its type.
| Sub-element (qualitative) | Description | # | R |
|---|---|---|---|
| methodID | A reference to a specific method that describes the context for the qualitative code. | 1 | Yes |
| code | The label of the category used for measurement values. | 1 | Yes |
| definition | Longer description of the definition of the category. | 0..1 | No |
| Sub-element (ordinal) | Description | # | R |
|---|---|---|---|
| methodID | A reference to a method that describes the context for the ordinal code. | 1 | Yes |
| code | Ordinal class code (e.g. a value like “+” or “1” in Braun-Blanquet cover scale) | 1 | Yes |
| definition | Longer description of the definition of the ordinal class. For example, “>1-5 % percent cover” for code “1” in an ordinal cover scale. | 0..1 | No |
| lowerLimit | Lower limit of the ordinal class in an associated quantitative scale (e.g. 10% cover in a cover class) | 0..1 | Yes |
| upperLimit | Upper limit of the ordinal class in an associated quantitative scale (e.g. 25% cover in a cover class) | 0..1 | Yes |
| order | Explicit order in the sequence of ordinal values to which this class belongs. | 0..1 | Yes |
| Sub-element (quantitative) | Description | # | R |
|---|---|---|---|
| methodID | A reference to a specific method that describes the context for the quantitative attribute. | 1 | Yes |
| unit | Unit of measurement (e.g. mm, cm, square meters, number of individuals). | 1 | Yes |
| precision | The smallest place value to which the measurement is expressed (eg, if pi is represented as 3.14, then its precision is .01). | 0..1 | No |
| lowerLimit | Potential lower limit of the measurement | 0..1 | Yes |
| upperLimit | Potential upper limit of the measurement | 0..1 | Yes |
Element organismName
An element organismName is simply a string with the name an organism used in the data set. This will normally be a nomenclatural unit of any rank (order, family, genus, species, subspecies, etc.). If it is a formal scientific name (not necessarily including authority) then the attribute ‘taxonName’ should be set to true. However, the organism name can be a morphospecies, a field name… cases in which the attribute ‘taxonName’ should be set to false.
| Attribute | Description | # | R |
|---|---|---|---|
| taxonName | A flag to identify that the organism name is a taxon name (i.e. a name according to a nomenclature code) | 1 | Yes |
Element taxonConcept
The representation of a taxon concept (i.e., an organism name and the organism description given by an author in a publication). A taxon concept may be referenced in an organism identity as the original concept used by the author of the data set, or it can be referenced in taxonDetermination allowing an organism identity to be mapped to a taxonomic concept by third parties after re-examination. Taxon concepts are uniquely determined by the organism (normally a taxon) name and a bibliographic citation.
| Sub-element (quantitative) | Description | # | R |
|---|---|---|---|
| organismNameID | A link to a specific organism name by ID. | 1 | Yes |
| accordingToCitationID | A link to a bibliographic reference by ID where the taxon concept is described. | 1 | Yes |
References
- Franz NM & Peet RK (2009). Towards a language for mapping relationships among taxonomic concepts. Systematics and Biodiversity 7: 5-20.
- Jones MB, Schildhauer MP, Reichman OJ & Bowers S (2006). The new bioinformatics: integrating ecological data from the gene to the biosphere. Annual Review of Ecology, Evolution and Systematics 37: 519-544.
- Wiser SK, Spencer N, De Caceres M, Kleikamp M, Boyle B & Peet RK (2011). Veg-X - an exchange standard for plot-based vegetation data. Journal of Vegetation Science 22: 598-609.